This project demonstrates the use of K-Means clustering and Singular Value Decomposition (SVD) to analyze the well-known Breast Cancer Wisconsin dataset using Apache Spark on Colab. The project includes various stages such as data preprocessing, clustering, dimensionality reduction, and comprehensive data visualization to illustrate the results.
🔍Setup
To run this project in Google Colab, first install the required packages and set up Spark and Java:
!pip install pyspark
!pip install -U -q PyDrive
!apt install openjdk-8-jdk-headless -qq
import os
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
🛠️Data Preprocessing
from sklearn.datasets import load_breast_cancer
breast_cancer = load_breast_cancer()
# Convert to Pandas and Spark DataFrames
pd_df = pd.DataFrame(breast_cancer.data, columns=breast_cancer.feature_names)
df = spark.createDataFrame(pd_df)
📊Clustering with K-Means
We apply K-Means clustering with k=2 (since the dataset has two classes: benign and malignant). The clustering performance is evaluated using the Silhouette score.
from pyspark.ml.clustering import KMeans
from pyspark.ml.evaluation import ClusteringEvaluator
kmeans = KMeans().setK(2).setSeed(1)
model = kmeans.fit(features)
predictions = model.transform(features)
# Calculate Silhouette score
evaluator = ClusteringEvaluator()
silhouette_score = evaluator.evaluate(predictions)
print(f'Silhouette Score: {silhouette_score}')
🔽Dimensionality Reduction
To optimize computational efficiency, we apply Singular Value Decomposition (SVD) to reduce the dimensionality of the dataset by a factor of 15x.
from pyspark.ml.feature import PCA
pca = PCA(k=2, inputCol="features", outputCol="svdFeatures")
pca_model = pca.fit(features)
svdFeatures = pca_model.transform(features).select("svdFeatures")
📈Results Comparison
- K-Means clustering was applied to classify the data into two clusters (Benign and Malignant), achieving a Silhouette Score of 0.834, demonstrating strong intra-cluster cohesion.
- Singular Value Decomposition (SVD) was used to reduce the dataset’s dimensionality by 15x while maintaining a Silhouette Score of 0.835, ensuring the model’s accuracy and efficiency post-reduction.
- The results confirmed that dimensionality reduction did not significantly impact clustering performance, while reducing computational costs.
# Perform K-Means on SVD-reduced data
kmeans_svd = KMeans().setK(2).setSeed(1).setFeaturesCol("svdFeatures")
model_svd = kmeans_svd.fit(svdFeatures)
# Silhouette score for reduced dataset
silhouette_score_svd = evaluator.evaluate(model_svd.transform(svdFeatures))
print(f'Silhouette Score (SVD): {silhouette_score_svd}')
📊Visualization
We include several visualizations to help understand the clustering and dimensionality reduction results:
1. PCA Visualization:
- A PCA plot shows the Breast Cancer dataset reduced to two components, visually displaying the separation between benign and malignant tumors.
from sklearn.decomposition import PCA
import matplotlib.pyplot as plt
pca = PCA(n_components=2)
pca_result = pca.fit_transform(breast_cancer.data)
plt.figure(figsize=(8, 6))
plt.scatter(pca_result[:, 0], pca_result[:, 1], c=breast_cancer.target, cmap='viridis', s=50)
plt.title("PCA of Breast Cancer Dataset")
plt.xlabel("PCA Component 1")
plt.ylabel("PCA Component 2")
plt.colorbar(label="Target (0: Benign, 1: Malignant)")
plt.show()
from sklearn.cluster import KMeans
kmeans = KMeans(n_clusters=2)
kmeans.fit(pca_result)
labels = kmeans.labels_
centroids = kmeans.cluster_centers_
plt.figure(figsize=(8, 6))
plt.scatter(pca_result[:, 0], pca_result[:, 1], c=labels, cmap='viridis', s=50)
plt.scatter(centroids[:, 0], centroids[:, 1], c='red', marker='X', s=200, label='Centroids')
plt.title("K-Means Clustering of Breast Cancer Dataset")
plt.xlabel("PCA Component 1")
plt.ylabel("PCA Component 2")
plt.legend()
plt.show()
3. SVD Clustering:
- A similar visualization is provided after applying SVD for dimensionality reduction, showing that the clusters remain well-separated even after reducing the dataset’s dimensionality by 15x.
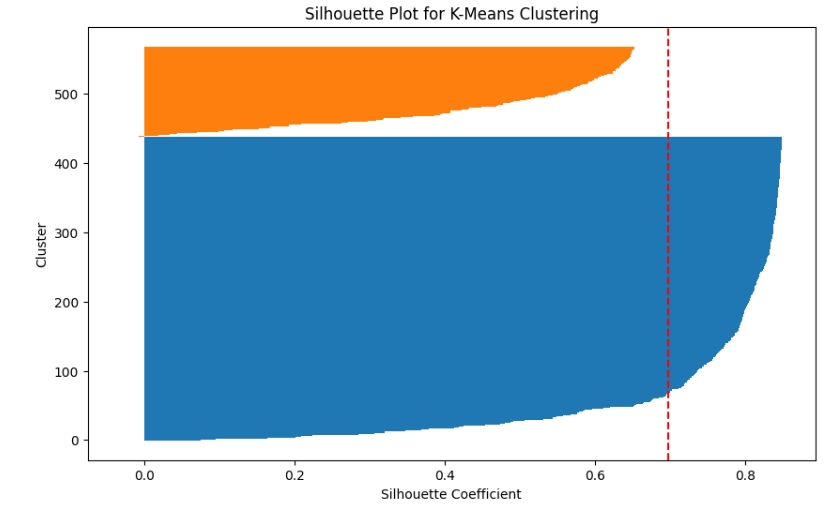
from sklearn.metrics import silhouette_samples, silhouette_score
silhouette_vals = silhouette_samples(breast_cancer.data, labels)
plt.figure(figsize=(10, 6))
for i in range(2):
cluster_silhouette_vals = silhouette_vals[labels == i]
cluster_silhouette_vals.sort()
plt.barh(range(len(cluster_silhouette_vals)), cluster_silhouette_vals, height=1.0)
plt.title("Silhouette Plot for K-Means Clustering")
plt.xlabel("Silhouette Coefficient")
plt.axvline(silhouette_score(breast_cancer.data, labels), color='red', linestyle='--')
plt.show()
📦Dependencies
pysparkscikit-learnpandasnumpy
🎯Conclusion
- K-Means clustering was applied to classify the data into two clusters (Benign and Malignant), achieving a Silhouette Score of 0.834.
- Singular Value Decomposition (SVD) reduced the dataset’s dimensionality by 15x while maintaining a Silhouette Score of 0.835.
- Dimensionality reduction optimized computational performance without sacrificing model accuracy.
🚀Running the Code
!pip install pyspark
!pip install -U -q PyDrive
!apt install openjdk-8-jdk-headless -qq